Improving Clinical Decision Support and Investigational Efforts with GenomOncology’s Drug Ontology
Key Takeaways
GenomOncology’s drug ontology includes FDA/EMA approved and investigational agents in oncology and is optimized for precision oncology use cases, including trial and therapy matching.
GenomOncology’s drug ontology can also be leveraged for research efforts, e.g. analyzing the landscape of approved and investigational therapeutics.
The ontology is expertly curated and continuously maintained.
Drugs’ mechanism of action (e.g. selective degrader), molecular target (e.g. EGFR targeting agent), modality (e.g. ADC), and other characteristics (e.g 2nd generation) are incorporated into the ontology.
Drugs have extensive synonym lists and database codes from sources like NCIt, JAX-CKB, OncoTree, SNOMED, ICD-9/10, and FDA.
Cancer is a complex and heterogeneous disease that often responds well to precision modalities of treatment. Pathologists and oncologists should have access to comprehensive and up-to-date information about the molecular characteristics of each patient’s tumor, as well as the available drugs and clinical trials that target those characteristics. Additionally, research scientists working in the development of oncology therapeutics or those designing clinical trials can also benefit from a deep understanding of both the existing and investigational therapeutic landscape.
GenomOncology’s (GO) drug ontology facilitates precision clinical decision-making and can support drug or trial development pipelines. Our ontology is a structured representation of the current knowledge on approved and developing oncology therapeutics, including graph-based representations of the modality, target, and biology of the agent. This extends to the indications and trials associated with a given drug. Additionally, our drug ontology helps organize, integrate, and standardize data from various sources- databases, prescribing labels, guidelines, and publications. Ultimately, the drug ontology primarily supports end-to-end tertiary analysis that can be accomplished with GO’s software solutions (e.g. clinical trial and therapy matching based on the unique molecular profile of a cancer patient), but it also can facilitate investigational efforts.
Development and Use of GO’s Drug Ontology
The drug ontology was bootstrapped from existing sources: mainly NCI Thesaurus (NCIt) and the JAX-Clinical Knowledge Base (JAX-CKB). NCIt is a reference terminology that covers various domains related to cancer research and care. JAX-CKB is a curated database that provides information on clinically relevant variants, therapies, mechanisms of action, clinical trials, etc. We perform weekly updates of the drug ontology using JAX-CKB’s update. Additional entities are ingested on an ad hoc basis to support the curation of clinical trials and therapeutic assertions (FDA Approvals, including accelerated development and review pipelines, Accelerated, Fast Track, Breakthrough Therapy, Priority Review, and Regenerative Medicine Advanced Therapy Designations, and guidelines from professional organizations like NCCN and ESMO). Together, this currently yields roughly 8,000 entries (7,869 as of 3/21/2023). GO’s In-House Content and Curation Team continuously maintains the ontology and all related elements.
As mentioned above, the drug ontology primarily supports three main active use cases: therapeutic assertions, clinical trial eligibility curation, and prior intervention criteria curation. Therapeutic assertions are structured statements that link a disease, setting, and biomarker data (e.g. genomic variants like BRAF V600E) with a specific therapy. These assertions are annotated with clinical guidance and evidence. We support both USA and EU therapies. Clinical trial eligibility curation involves extracting relevant eligibility criteria (e.g. biomarkers, diseases, disease states, treatment settings, prior intervention requirements) from trial protocols. These criteria are then mapped to structured ontologies and connected with logical operators so the eligibility logical statements can be used in our trial match algorithm. Within trial curation, we build “Treatment Contexts,” which can be thought of as a given treatment arm of a trial. In Treatment Context, we curate the drug(s) and setting(s), and this is then linked to the disease eligibility group on the trial. We also build prior intervention criteria curation; we analyze trial documents and curate inclusion or exclusion statements to do with prior therapy. For example, a trial may be testing a new HER2 targeting drug and only want patients who have progressed on already approved anti-HER2 therapies like trastuzumab. We can capture and structure this important data related to trial eligibility because of the drug ontology. All these use cases for the drug ontology enables patient matching.
Additional Advantages and Uses for GO’s Drug Ontology
Other drug ontologies and databases relevant to precision oncology of course exist, but our ontology has several characteristics that make it optimal for precision oncology use cases and investigational research. GO’s API suite enables client-specific customization and enhances portability, making integration of our data into third party applications and workflows simple. And because our ontology is designed to be cancer-focused, we have thoughtfully minimized non-cancer related agents and prioritized oncologic agents. Additionally, the ontology is expertly curated ensuring data rich, multidimensional entries. This makes filtering, querying, and operationalizing our drug data straightforward. Plus, the ontology is continuously maintained and updated by our team of experts, who closely track the investigational landscape. New agents, annotations and drug-related findings are incorporated weekly. Altogether, the special features of our drug ontology can benefit both clinicians and investigators in the precision oncology space. It is a rich and robust resource that can help answer complex questions.
Structure
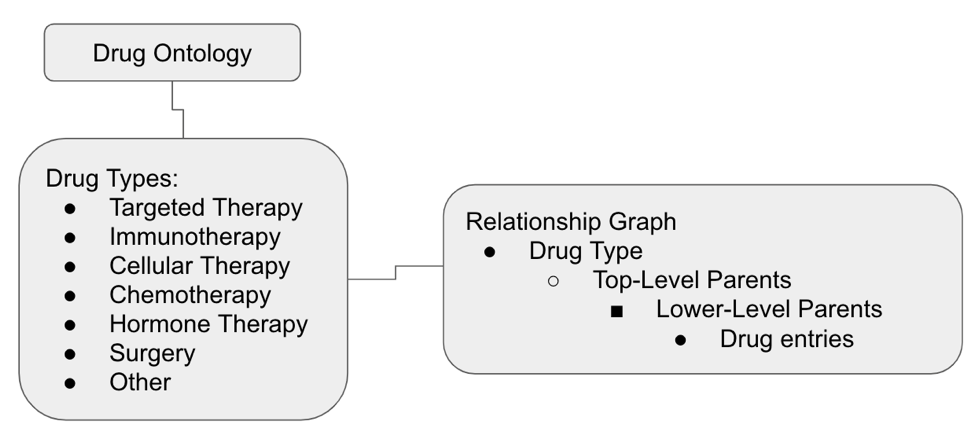
The drug ontology has a hierarchical structure with seven top-level categories, which we call “Drug Types:” Targeted Therapy, Immunotherapy, Cellular Therapy, Chemotherapy, Hormone Therapy, Surgery, and Other. Within each Drug Type, drugs are organized by various “Parents” or subcategories that reflect different aspects of drugs, such as mechanism of action, molecular target, or modality.
The ontology is graph-based, meaning there can be multiple parental or daughter relations for any entry and cases of drugs that blend Drug Types can be handled without breaking our matching logic. This approach allows for flexibility and expressivity in representing complex relationships among drugs.
Our entries have extensive synonym lists and database codes from sources like NCIt, JAX-CKB, OncoTree, SNOMED, ICD-9/10, and FDA. This helps with harmonizing different terminologies and facilitating interoperability.
Drug Ontology Insights
Beyond the use previously described, additional insights can be derived using the GO drug ontology during tertiary analysis of somatic cancer profiles. It can…
reduce ambiguity and inconsistency in naming and classifying drugs across different sources and platforms.
improve accuracy and completeness in identifying relevant drugs and trials for each patient’s tumor profile.
enhance efficiency and scalability in curating and updating information about drugs and trials using automated methods.
enable advanced analytics and decision support tools based on semantic reasoning and inference over the drug knowledge base.
GO’s drug ontology can also support drug development pipelines or help with the design of clinical trials in several ways:
provide a comprehensive and standardized vocabulary for describing drug targets, mechanisms of action, indications, contraindications, and outcomes.
enable data integration and interoperability across different sources and formats of drug discovery data by using common identifiers and semantic annotations.
facilitate data analysis and knowledge discovery by supporting complex queries and reasoning over drug-related information using natural language or graphical interfaces.
To learn more, request more information today.